Our research. Our methods.
-
![]()
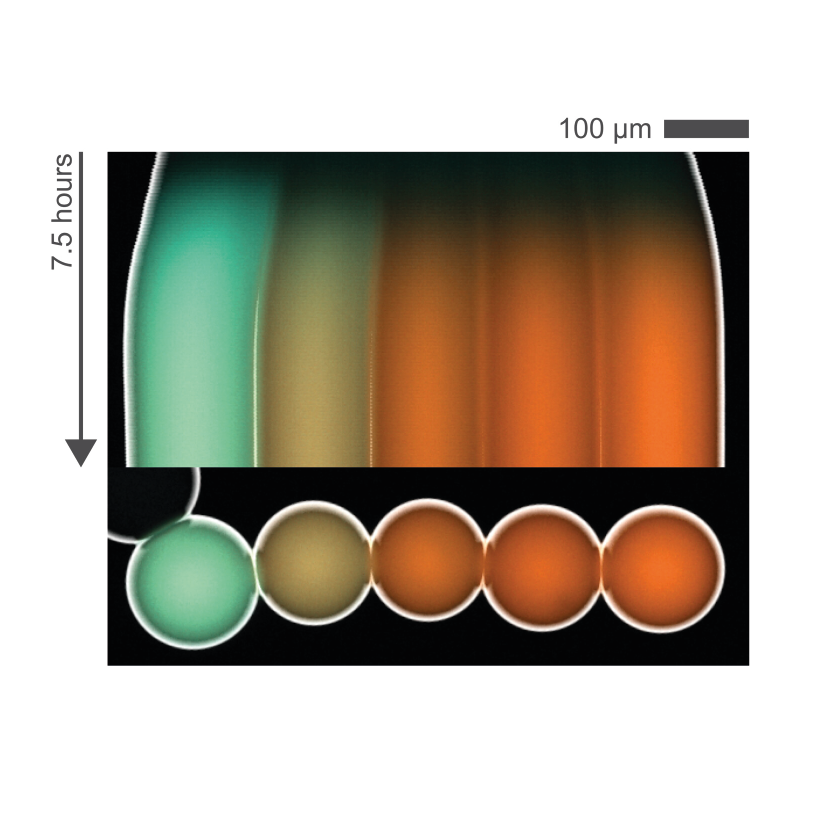
Cell-free gene regulation
Living organisms have fascinating abilities to sense and adjust to perturbations in their environment. They regulate their chemical, mechanical, metabolic responses by activating and deactivating genes dynamically.
Examples that inspire us include pathogen detection by the immune system, robust embryonic patterning, and coordinated responses in populations of cells.
We want to understand the general principles of this regulation beyond in vivo specificities. We do this by programming genes in cell-free environments and challenging them in wide ranges of control parameters. We build synthetic gene circuits to test how different regulation mechanisms respond in space and time.
-
![]()
Synthetic immunology
The immune system’s capacity to tell what is foreign from what is us is one of the most fascinating function of living systems, both from a biological and from a philosophical stand point.
How is such a global response as immunity programmed through molecular protein-protein interactions? How does our immune system detect pathogens while passing over our own proteins, even when they look very similar? And what can make our immune proteins fail at making this distinction, so they attack ourselves or become blind to threats?
We use a synthetic biology approach to look at these mechanisms of immune recognition from the bottom-up. We express immune proteins, like antibodies and antigens, and we explore how they interact, and what disturbs these interactions. This unique approach is part of synthetic immunology, an emerging field that tackles fundamental immunology questions using engineering tools.
-
![]()
Methodologies
As synthetic biologists, we take an engineering approach to studying life. Our methods allow us to build plug-and-play platforms where DNA and proteins can be programmed in space and time.
Cell-extract: We extract the contents of E.coli cells so that we can express synthetic DNA into proteins in our platform. Cell-extract preparation and composition determines the dynamics of protein expression.
Biochips: We use a unique approach developed by the Bar-Ziv lab to model cells while having full control over our platforms. We carve microcompartments on silicon chips that are cell-sized. Inside these compartments, DNA and proteins can be attached to surfaces, so that genotype and phenotype are colocalized while keeping the advantages of an open accessible environment.
Fluorescent microscopy: We use fluorescent reporters, such as GFP and RNA aptamers, to measure the dynamics of gene response at the transcription and translation levels, through time and space.
Gene circuits: We design positive and negative feedbacks between genes to encode regulation, response to signals, differentiation, and other biological functions.